Introduction
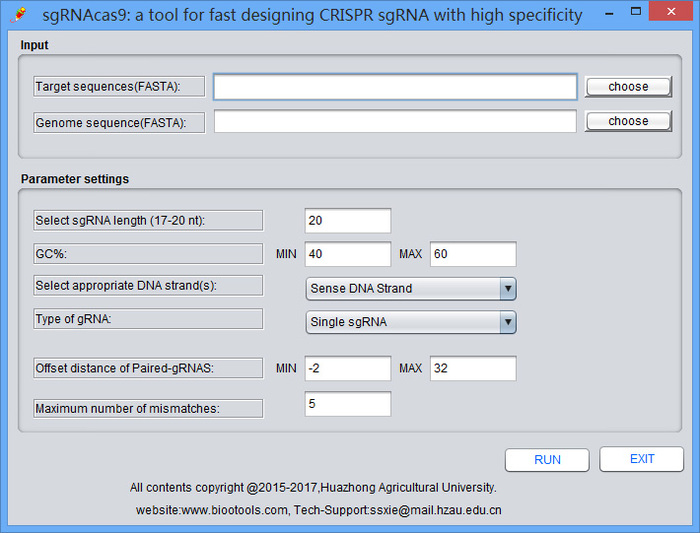
A software package developed for fast design of CRISPR sgRNA with minimized off-target effect, in order to improve CRISPR-Cas9 specificity for targeted genome editing and transcriptional control. This package consists of programs to perform a search for CRISPR target sites (protospacers) with user-defined parameters, predict genome-wide Cas9 potential off-target cleavage sites (POT), classify the POT into three categories, batch design oligonucleotides for constructing 20-nt (nucleotides) or truncated sgRNA expression vectors, extract desired length nucleotide sequences flanking the on- or off-target cleavage sites for designing PCR primer pairs to validate the mutations by T7ENI cleavage assay. Importantly, by identifying potential off-target sites in silico, the sgRNAcas9 allows the selection of more specific target sites and aids the identification of bona fide off-target sites, significantly facilitating the design of sgRNA for genome editing applications. In July, 2015, the sgRNAcas9 (GUI) software v2.0, developed independently by the Huazhong Agricultural University, was certified by china state bureau of copyrights (No. 2015SR170234). sgRNAcas9 sgRNA design source code free for academic, nonprofit, and personal use.
Downloads
If you use this program in your research, please cite:
Xie S, Shen B, Zhang C, Huang X, Zhang Y.sgRNAcas9: a software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites. PLoS One. 2014 Jun 23;9(6):e100448. doi: 10.1371/journal.pone.0100448. eCollection 2014. (link)
Introduction
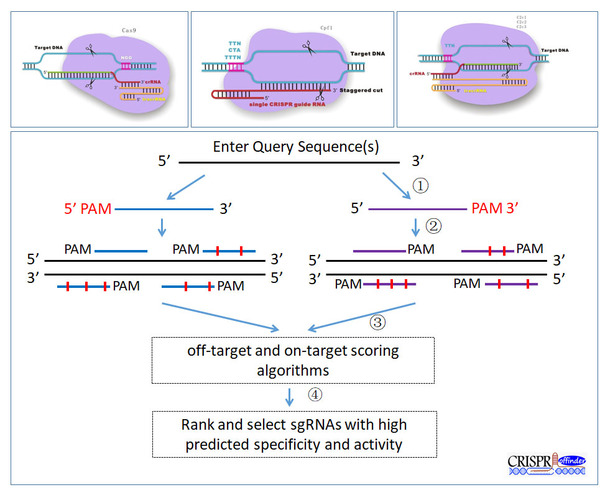
A flexible sgRNA design program named “CRISPR-offinder” was developed. Support for user-defined PAM and sgRNA length was provided to increase the targeting range and specificity. Additionally, evaluation of on- and off-target scoring algorithms was integrated into the CRISPR-offinder. The CRISPR-offinder has provided the bench biologist a rapid and efficient tool for identification of high quality target sites.
Donwloads
User_manual_CRISPR-offinder.pdf
If you use this program in your research, please cite:
Zhao C, Zheng X, Qu W, Li G, Li X, Miao YL, Han X, Liu X, Li Z, Ma Y, Shao Q, Li H, Sun F, Xie S, Zhao S. CRISPR-offinder: a CRISPR guide RNA design and off-target searching tool for user-defined protospacer adjacent motif. Int J Biol Sci. 2017 Nov 1;13(12):1470-1478. doi: 10.7150/ijbs.21312. eCollection 2017. (link)
Introduction
CRISPRamplicon, an analysis tool that unites identification, quantification and visualization of genuine genome editing events from High-throughput sequencing data, for example, CRISPR amplicon sequencing data, Target capture sequencing data, Whole exome sequencing data, and Whole genome sequencing data, etc.
Downloads
If you use this program in your research, please cite:
Zhao et al., Effect of sequence length and microsatellite instability on sgRNA activity and specificity. 2019, In prepare
Introduction
CRISPR-lib is a command line tool, tailored for fast end-to-end design of sgRNA libraries for any species.
Downloads:
Coming soon!
If you use this program in your research, please cite:
Xie et al., CRISPR-lib: a local single guide RNA libraries design tool for multispecies, 2019, In prepare.
Introduction
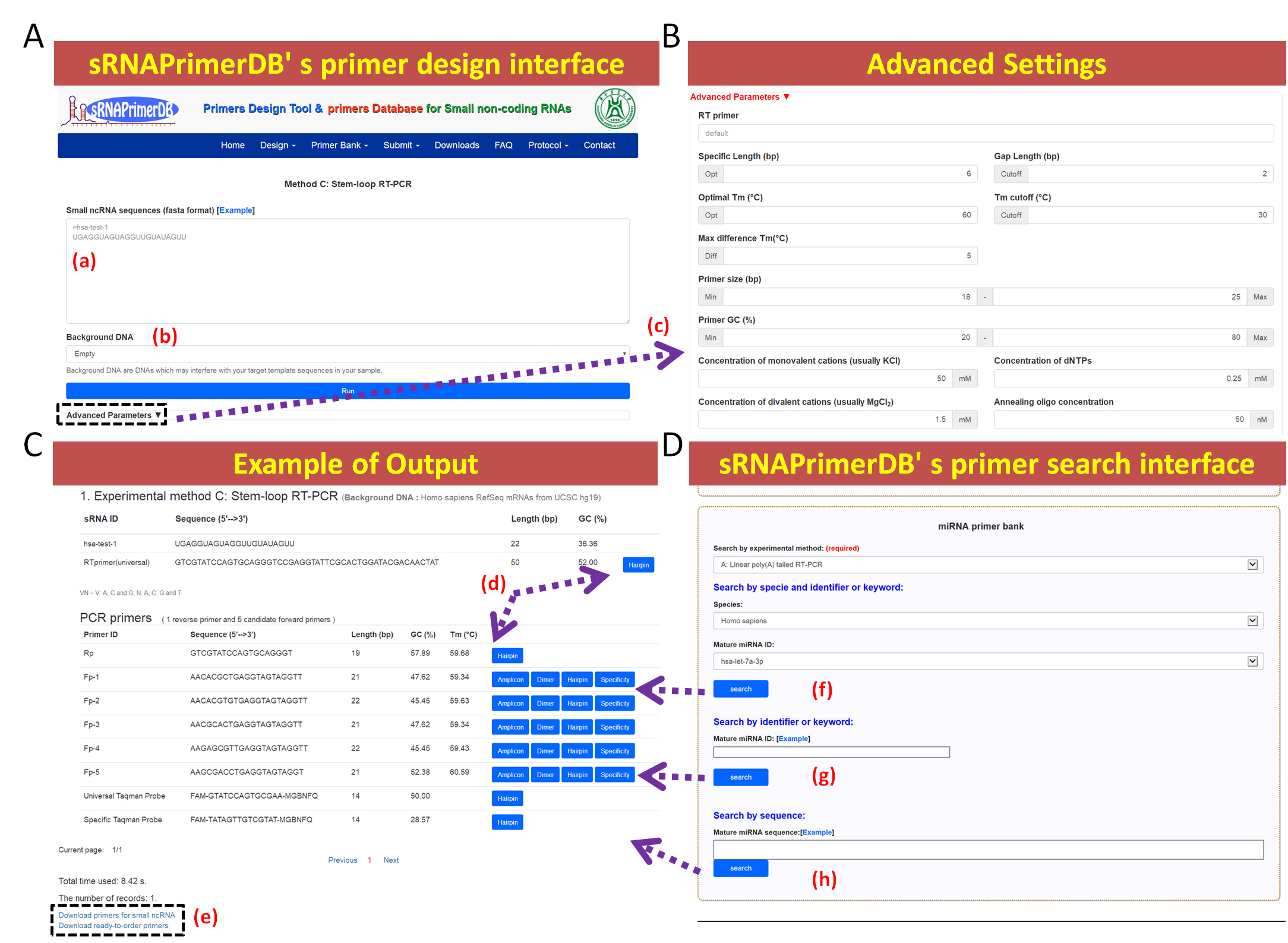
sRNAPrimerDB is composed of multiple primer designing programs, combined with calculated primers. It allows users to search and design primers for small ncRNAs, such as miRNAs and piRNAs. The database is composed of eight divisions including a section for designing, a bank of pre-designed primers and probes, a searching tool as well as links towards scientific literature and resources. Primers for small ncRNAs that have been experimentally validated are encouraged to submit an online application form by visiting our web site at www.srnaprimerdb.com.
If you use this program in your research, please cite:
Xie S#, Zhu Q#, Qu W#, Xu Z, Liu X, Li X, Li S, Ma W, Miao Y, Zhang L, Du X, Dong W, Li H, Zhao C, Wang Y, Fang Y*, Zhao S*. sRNAPrimerDB: Comprehensive primer design and search web service for small non-coding RNAs.Bioinformatics. 2018 Oct 8. doi: 10.1093/bioinformatics/bty852.